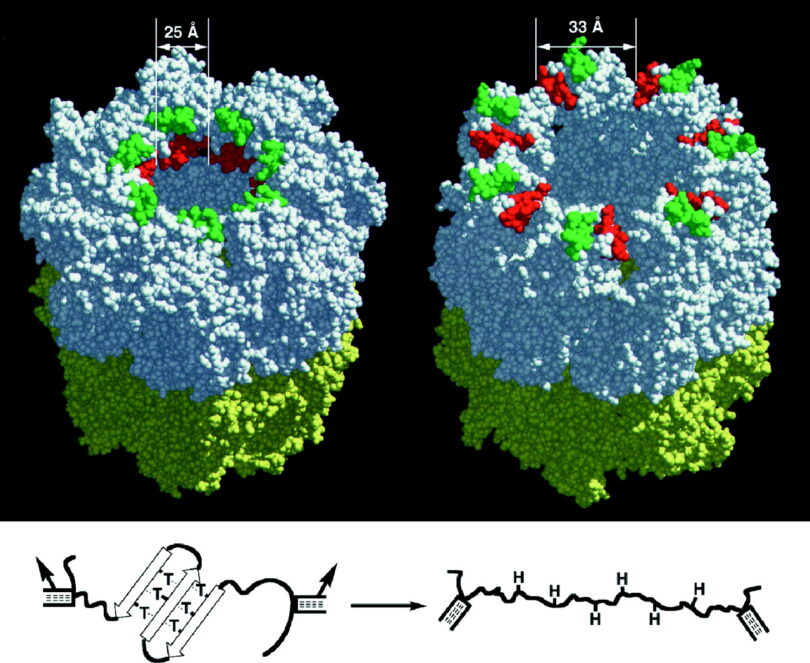
Chaperonin’s: Groups I & II
Chaperonins operate as nanomolecular machines inside the living cell. These proteins form into specialized barrels that take an unfolded protein, encapsulate it, and assist in its final properly folded three-dimensional shape. There are three types of chaperonins, creatively and imaginatively named Groups I, II, & III. Looking at the image below, the top of these protein machines open and reveal a portal where an unfolded protein enters (see C & D as the top view). After the protein enters the chaperonin, the top closes, and the protein inside is folded. When the folding is complete, the top of the barrel-shaped chaperonin re-opens and releases the folded protein for use in the living organism.
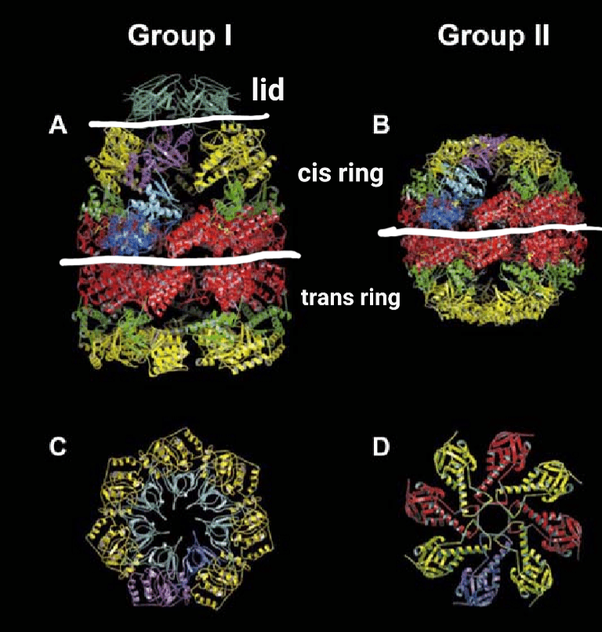
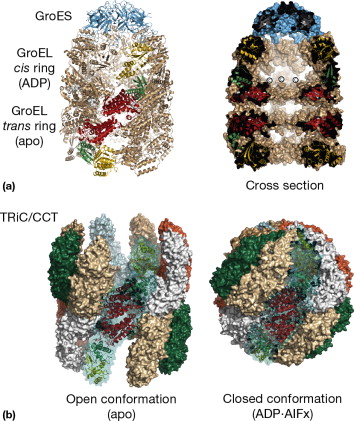
Notice how remarkably similar the structures between Group I and Group II are (within the white lines drawn onto the image). These chaperonin Groups are composed of seven identical proteins that bind together to make a hollow donut shape called the “cis ring.” Another ring of the same seven proteins is oriented oppositely and stacked below the cis ring, which creates an empty barrel for an unfolded protein to go inside.
Unique to Group I is the lid. Group II has eight different proteins that make the cis ring, and in like fashion, a second identical oppositely orientated ring stacks together, making an empty barrel. Group II has no lid, as the cis and trans rings change shapes to open and close. We will get to Group III below.
Given the uncanny functional and structural similarities between these protein machines, one would assume that Group II evolved from Group I. That’s what evolutionary biologists will tell you. First, there is no significant sequence similarity that supports either genetically. ¹ Second, the small number of alignable amino acid positions between the Group I and the Group II chaperonins provide little phylogenetic signal to address the supposed evolutionary history of the archaeal/eukaryotic chaperonin tree.²
In other words, there is no DNA sequence similarity between the Group I and II chaperonins, yet we are assured that Group II must have evolved from Group I. If not, this would not reflect kindly on expectations of universal common descent. We cannot have two stunningly similar systems doing similar functions within the cell, with genetic sequences largely independent. This is the opposite of what Darwinism would expect, yet this is precisely what we observe.
The sequences between Group I and Group II are widely dissimilar genetically. Therefore, if Group II didn’t “evolve” from Group I, then how can evolution explain its origin? Did Group II just poof into existence all on its own?
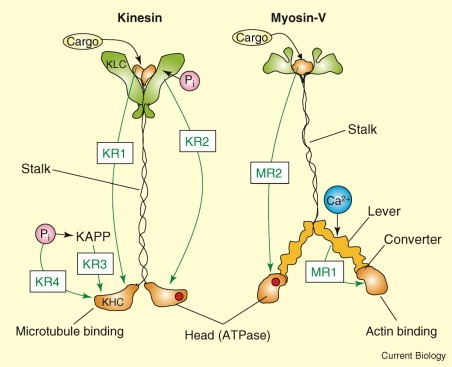
This type of scenario is ridiculously common in life. The protein skeletons inside the cells of prokaryotes are functionally and structurally the same as the protein skeletons inside of eukaryotes, but they are not sequence-similar. Protein motors like Kinesin and Myosin are other examples of structurally and functionally similar machines without sequence similarity. So-called “orphan genes” are those sequences fully unique to each life form, each offering no evidence of common descent. The name “orphan” is the conclusion- not the observation.
Chaperonin’s: Group III
Group III, like Group II, has two concentric rings composed of 8 different proteins. Also, it does not have a lid like Group I but the specialized rings function as a sophisticated portal for both opening and closing the protein chain for folding.
Phylogenetic analyses of Chaperonin’s betray common descent expectations
These DNA comparisons show that Group III must have evolved from Group II and not Group I.³ This indicates that Group III was the last of the three types of chaperonins to arrive. This presents another problem for Darwinism since Group I & III are found in Bacteria, but not in Group II. Bacteria are prokaryotes that allegedly precede Archaea and eukaryotes in the evolutionary timeline.⁴ Yet, Group II does not show up until Archaea. How, then are Group III present in Bacteria if they evolved from Group II?
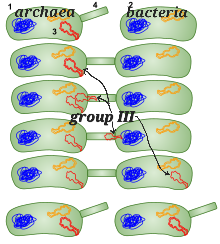
Evolutionary biologists claim that Bacteria first evolved Group I, and later Archaea evolved Group II, through which Group III evolved. Then Archaea squirted the genes for Group III into Bacteria in a process called lateral gene transfer. The image explains this hypothesis that Archaea (on the left) inserted DNA into Bacteria (giving it Group III chaperonins).
Here’s why this is a failed attempt to explain away the evidence. The DNA sequence (gene) that codes for Group III chaperonins in Bacteria is found in an “operon,” or a cluster of genes. The following is the operon.⁵ It’s a string of DNA with each arrow representing a specific gene (the name is above the arrows).
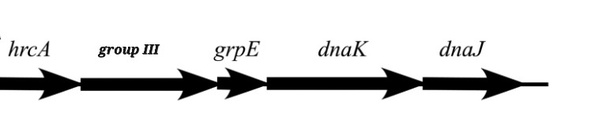
If Archaea transferred Group III to Bacteria, which is the operon we find in all Bacteria that possess Group III, this operon should be present in Archaea, but it’s not… because, we are told, lateral gene transfer did not happen here.
Secondly, the dnaK gene found in Archaea is in its own family relative to the one found in Bacteria with Group III.⁶ If the operon containing dnaK and Group III in Bacteria came from Archaea, both Bacteria and Archaea should have similar dnaK genes… but they don’t.
Lastly, all Bacteria code for their own dnaK genes, so if this operon were given to Bacteria from Archaea, then the Bacteria possessing Group III would have multiple copies of the dnaK gene, but they do not because lateral gene transfer didn’t happen here. How can Archaea pass to Bacteria what they themselves do not possess? It cannot provide genetic sequences it does not possess.
Conclusions
Based on genetic sequence similarities, Group II did not evolve from Group I. Group III did not evolve from Group II, as indicated by its presence in Bacteria. The evidence clearly shows that similar nanomachines with similar biological functions do not have sequence similarity. Darwinian theory expects to find similarities in genetics to describe the transmutation of species from Bacteria to Humans. Here, we find massively complex structures comprised of multiple proteins that perform highly specialized protein folding within living things, but they have dissimilarity, and there are gaps in their supposed chronological order of metamorphosis. Does this disprove Darwinism? No, but it is a puzzle.

The coding regions of DNA (representing only about 1.2% of its total volume) between Humans and Bananas are up to 50% similar.7
Similarities of genetic sequences, anatomy, and physiological function within life (and structural similarities in fossil forms) are commonly used as an argument for the Darwinian Theory. However, similarity can just as easily be argued for an intelligent designer. We routinely find that information, order, function, and purpose are design attributes. From all we have ever observed, outside the claims of biology, all design only emerges from an intelligent mind. There are no exceptions. Richard Dawkins said that biology was the study of complex things that appear to be designed- but weren’t. The design implications within biology are devastating the Darwinism.
Does similarity implicate design or randomness?
However, even if chaperonins, protein skeletons, protein motors like Kinesin and Myosin, or orphan genes were all sequentially similar, which they are not, this would not button up the claims of Darwin’s theory. While we find structurally and functionally similar biological machines without sequence similarity, we also find many gene sequences that code for similar proteins used in similar environments (lungs on land, gils in water), but this also fails to infer universal common descent evolution strongly. Sure, sequence and anatomical similarities can rightly infer universal common descent; however, on the flip side of this argument, the incredible informational storehouses, order, and functional purposes behind such similarities also strongly infer an intelligent designer.
1- https://onlinelibrary.wiley.com/doi/10.1002/prot.22952
2- Origin and Evolution of Eukaryotic Chaperonins
3- Two chaperonin systems in bacterial genomes with distinct ecological roles – PubMed
5- https://www.pnas.org/doi/full/10.1073/pnas.1004783107#core-B40
7- “Are We Genetically Similar To Bananas And Why Is This Important For Research In Disease?” Sano, Nov, 2019; https://sanogenetics.com/blog/are-we-genetically-similar-to-bananas-and-why-is-this-important-for-research-in-disease/