From: http://www.dstoner.net/Math_Science/cell1.html
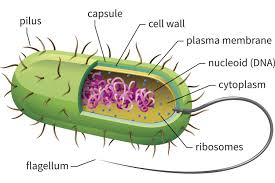
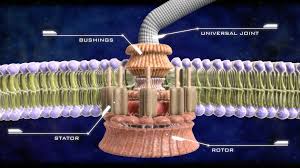
Prokaryotes are the “simplest” known living organisms. Bacteria flagellum are such prokaryotes. This overview is grossly over simplified but it is designed to reveal the extremely complex and nano precision and cascade of events required for prokaryotes cellular function. Many functions are left out of this overview and many functions remain well beyond the grasp of human understanding at this time in science.
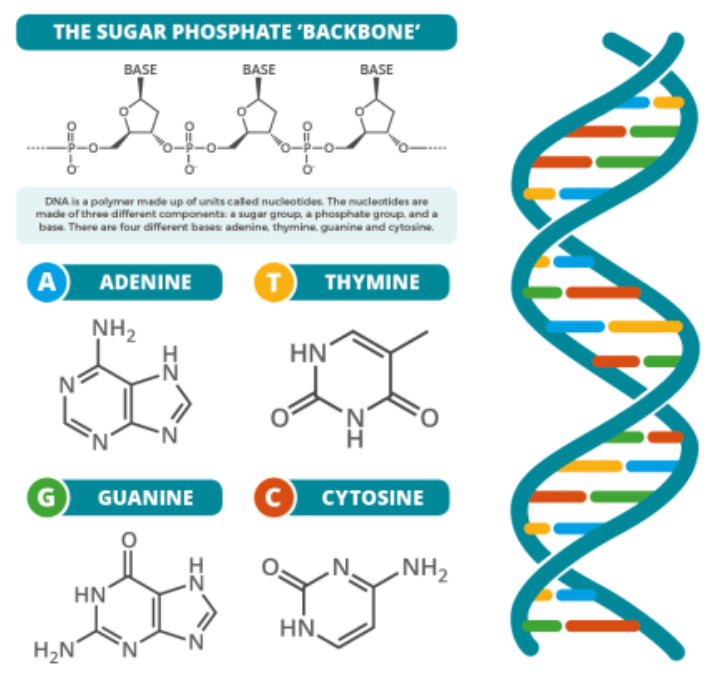
DNA: Is a base-4 information storage molecule, comprised of phosphates (sugars) and four chemical code constructs abbreviated as A, T, G, and C — paired A-T and G-C. The structure in a double helix much like rungs in a flexible ladder. Every three of these pairs (43 or 64 possible combinations) specifies either one of 20 amino acid molecules to make proteins.
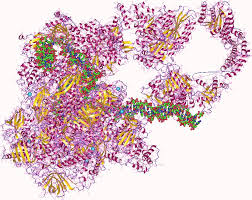
RNA Polymerase molecule: RNA Polymerase, with the help of many other special molecules, scans the DNA until it finds a legal start sequence, attaches itself, and “crawls” along the chain matching bits of RNA (similar to DNA but different in a few critical points) to the pattern on the DNA, connecting them together, and thereby constructing very specific RNA chains (specified by the exact pattern of the DNA molecule. These chains fold into complex and specific shapes (DNA doesn’t fold like this), some of which are themselves little machines. Three types of these produced RNA molecules are m-RNA, t-RNA, and r-RNA.
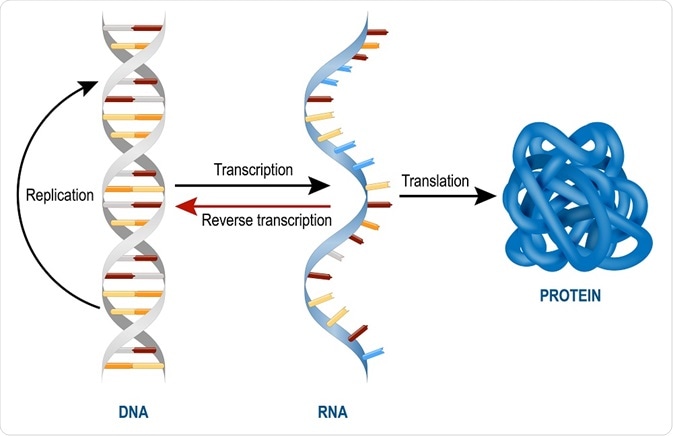
The Messenger RNA molecule: m-RNA is, like DNA, a data carrier — except that it doesn’t replicate or do many other things the DNA molecule does. Unlike DNA’s information, RNA’s information is easily accessible (but also more subject to damage). It is short-lived but can be read by another piece of cellular machinery; specifically the r-RNA molecule.
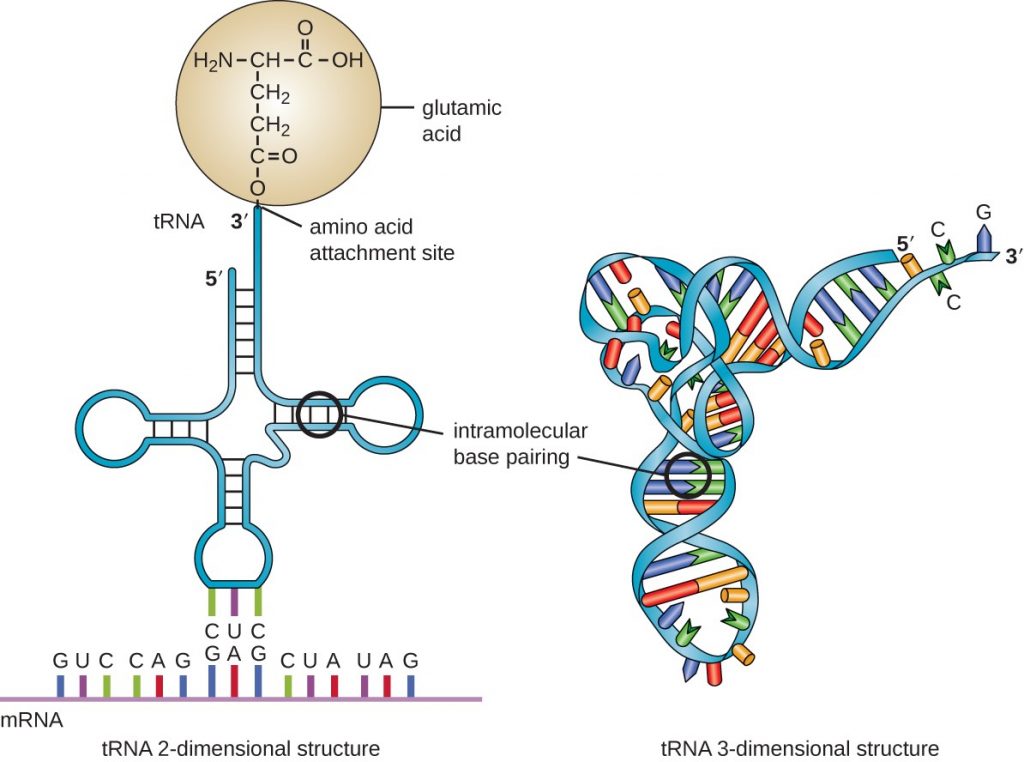
The Transfer RNA molecules: t-RNA hunts around the cell for specific amino acids. There is a different t-RNA for one of each of the 20 different amino acids used by life. Each t-RNA is also coded with three RNA markers (kind of like a bar code) also called a “codon” which identifies what amino acid it carries so that the ribosome can determine what kind of amino acid it is and thereby assemble to protein chain to the recipe.
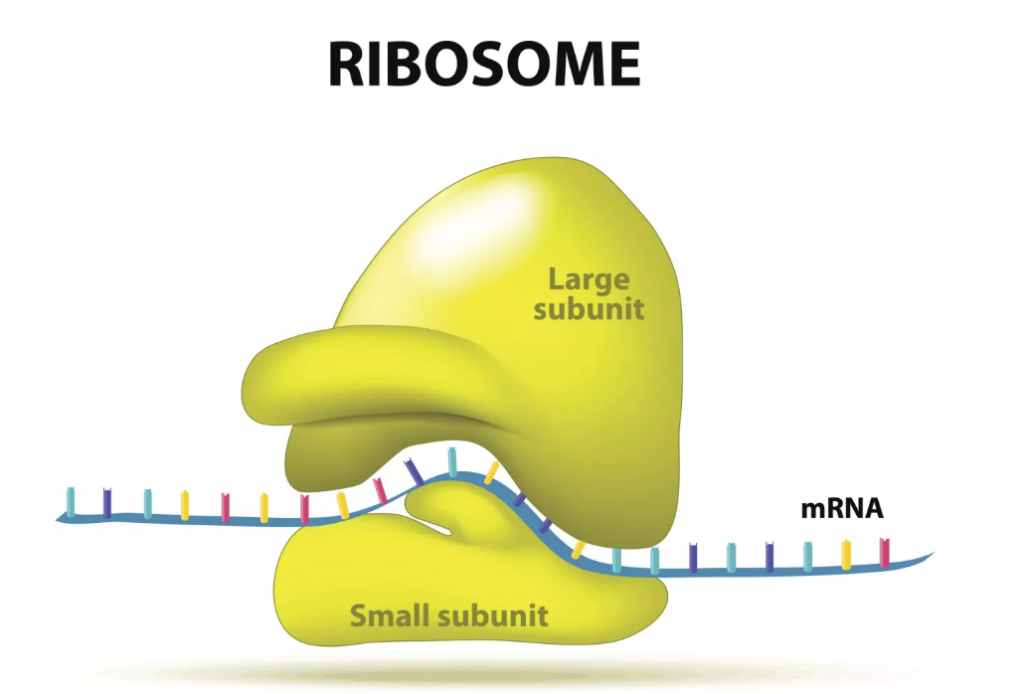
The Ribosomes r-RNA: Ribosomes, whose actual structure includes more than fifty different proteins in addition to their RNA, are tiny nano-tech numerically-controlled molecular assembly machines. They take individual amino acids (sorted and delivered to them by t-RNA molecules), select the correct ones (using the pattern supplied by m-RNA molecules and the “bar codes” on the t-RNA), align them (using little alignment jigs), then connect them together into a chain (while driving away water molecules, that would inhibit chemical bonding, by hitting them with tiny little hammers that fly back and forth at about 10 or 20 times a second), and finally check for errors (discarding incorrectly assembled amino-acid protein chains).
These tiny machines are powered by ATP molecules which prokaryotes produce directly in their cell wall and cytosol. In other types of cells (called eukaryotes), this ATP is produced in a different piece of chemical machinery called mitochondria (which have there own slightly different DNA and genetic code). Prokaryotes do not have mitochondria. This is how Ribosomes produce the many different kinds of proteins, including RNA Polymerase (see #2 above) and the 50 or so different proteins which they themselves require.
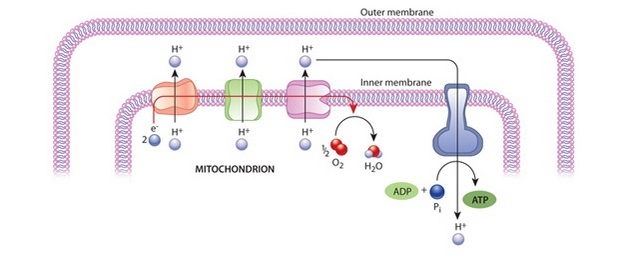
In conclusion, all very simple really.